RefNote: The evolution of multiple active site configurations in a designed enzyme KE07 & Evolutionary Optimization of Computationally Designed Enzymes: Kemp Eliminases of the KE07 Series
Evolutionary Optimization of Computationally Designed Enzymes: Kemp Eliminases of the KE07 Series
https://www.sciencedirect.com/science/article/pii/S0022283609015381?via%3Dihub
The evolution of multiple active site configurations in a designed enzyme
https://www.nature.com/articles/s41467-018-06305-y
(JMB 2010) Evolutionary Optimization of Computationally Designed Enzymes: Kemp Eliminases of the KE07 Series
This work did following to understand the KE07 DE process:
- crystal structural based analysis
- pH optimum experiment
Evolution change of KE07
- The salt bridge between E101 and K222 influence the pKa of E101
- I7D break this salt bridge to enhance the catalysis
- D7I in R7 cause kcat/Km decrease
- pH optimum after the mutation increased to ~6 from <4.5
- K222 in R7 is important
- K222A in R7 cause 500-fold kcat/Km decrease
- K222A in KE07 cause ~3-fold kcat/Km increase
- didn’t give any reason that make sense
- I7D break this salt bridge to enhance the catalysis
- The H-bonding network in 201-202-224 make H201 in a position of stabilizing TS
- N224D form H-bond with R202 and H201 that make them closer to the substrate
- the relationship between closer H201 and lower TS - R is not well explained
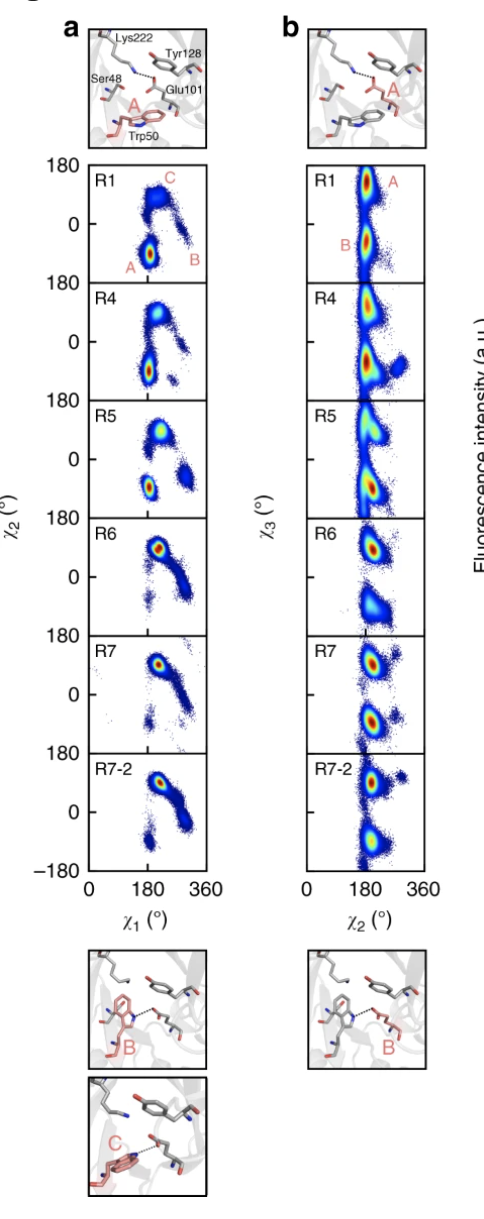
(Nat. Comm.) The evolution of multiple active site configurations in a designed enzyme
This work did following to understand the KE07 DE process:
- crystal structural based analysis
- KIE kinetics (how did they used that)
- Arrhenius kinetics in 8 temperature
- PROPKA calculation of E101
- fluorescence spectroscopy pf W50 (to probe the negative charge density on E101)
- HREX-MD enhanced sampling (for E101 conformation, modelled w/o ligand)
- EVB calcualtion of the reaction barrier of the 3 configs.
Mechanism of KE07
- If the pH value goes too high (too basic), the HIS tag will be deprotonated and bind into the pocket.
- After R4, present of product will break K222 salt bridge and form H bond with oxyanion of the product
- W50A signicantly reduce kcat
Evolution change of KE07
- R4: significantly lower Ea
- I7D lead to breaking of the salt bridge: better electrostatics but worse dynamics on E101 conformer (see the JMB paper)
- R5&R6: mainly the entropy part. Ea even increased from R5 to R6. (So it is actually reasonable)
- (???) based on 6DNJ they think R5 developed an additional catalytically competent conformation. (but it seems not a valid structure)
- And they can only observe conf. B when HIS tag is binding
- But HREX-MD sampled it and EVB suggests it works.
- really developed a new conformation C of W50 and is dominated. Conf. C is forming H-bond with E101 which pre-organize it for catalysis
- which is lower is activation barrier in this conformation comparing to A. (Prob due to organize E101)
- (???) based on 6DNJ they think R5 developed an additional catalytically competent conformation. (but it seems not a valid structure)
NOTE
studies related to KE07-R7-2 should use 6CT3 as the starting structure. (Use config )