RefNote: Experimental Thermodynamic data of Enzyme Interaction
Misc about experimental thermodynamic data of enzyme interactions
Method: Isothermal Titration Calorimetry (ITC)
Experiment
Isothermal
- Titration drop in Sample Cell cause temperature change
- Reference Cell at given constant temperature.
- The feedback system transfer energy in/out the Sample Cell to keep the temperature constant after every drop.
- The monitor record the energy transfer as peak.
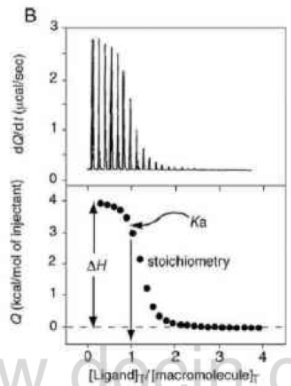
Data Analysis
Enthalpy
Integration of peak area directly gives the enthalpy($\Delta H$).
Entropy
$\Delta G = -RTlnK$
$\Delta G = \Delta H - T\Delta S$
Modeling the curve around the end point gives the K.
Dilution is required when the curve is too sharp to model.
Data: Different System
1BRS monomer
ITC
- Barnase to Barstar (pH=8)
- No aggression
- One to One “independent of protein concentration or whether barnase is titrated with barstar or vice versa”
- Preventing the aggression and dimerization. “24 mM Hepes buffer (pH 8), 1 mM DTT because barstar tends to aggregate in the cell of the calorimeter in Tris buffer at pH 8.The DTT was added to the buffer to prevent oxidation and dimerization of barstar.”
- IBRS-2 is not the correct reference (Not the ITC method) The 1BRS-2 paper(Fersht,1995) used the Tris buffer.
CRYSTAL
- Sequence
C40A C82A
Double mutant due to possilbe RedOx.
“Crystallographic studies involving wild-type barstar have been thwarted due to the unavoidable presence of a mixture of both oxidized and reduced species, thus necessitating the use of the double Cys—>Ala mutant. “ - Polymerism
Not really a trimer in the PDB, just the asymmetric section. / Chain AD was used in the analysis of the original reference
many problems in this part of PDB database(a polymer or monomers):Global Stoichiometryvalue to show what the author intended to get. Fits other data generation experiments for the most cases.(In solvent)- But this value cannot always indicate which chains is the target result. (e.g. 1NTO)
- Quality
- Differences within the Unit
Chain C have much higher overall temperature factor.
“This is most likely due to a lack of stabilizing crystal packing around this molecule, which is also less solvated than the other molecules in the asymmetric unit” - Exceptions
Missing barnase residues 1 and 2 from the N-terminus. (A,C)
This N-terminal disorder is a common feature in all barnase structures solved to date
These residues are present, however, in the barnase B chain of the barnase-barstar complex as a result of stabilizing crystal packing interactions
Missing barstar residue 1 from the N-terminus. (E)
There is no density for the N-terminal lysine residue of barstar chain E.
Missing barstar residue 64-65. (E,D)
It was impossible to build in residues 64 and 65 of barstar chains D and E because of relatively high disorder in this region of barstar.
- Differences within the Unit
MD
- Model
- Use Chain B for barnase. (Fully Modeled)
- Use Chain F for barstar. (Partially Modeled)
Fixed with Chain E. (K22 E28 S89)
Fixed with tLEaP. (E64 N65)
Mutate back to WT with the MutaGen.
1JTG monomer
ITC
- TEM-1-beta-lactamase to BLIP (pH=7) “titrating 40 M TEM-1-beta-lactamase into 4 M BLIP mutant in PBS (50 mM phosphate, pH 7.0, 150 mM NaCl).”
- Minimal proton linkage effect The binding enthalpy in Tris buffer was identical to that in phosphate buffer
CRYSTAL
- V59I A159V
1LFD
ITC
- ???